Research
The EnBiSys Lab is a highly collaborative, multidisciplinary research group housed in the Department of Electrical and Computer Engineering and the N.C. Plant Sciences Initiative at NC State University. We collaborate with researchers in many diverse fields, such as plant and microbial biology, forestry, chemical engineering and others. We develop techniques for understanding and influencing biological systems by using theories and methodologies from traditional electrical engineering areas, such as:
- Machine Learning
- Digital and Biological Signal and Data Processing
- Control Theory
- Computer Vision and Image Processing
- Nonlinear Systems Analysis
Current Projects
Improving Crop Productivity and Value Through Heterogeneous Data Integration, Analytics and Decision Support Platforms
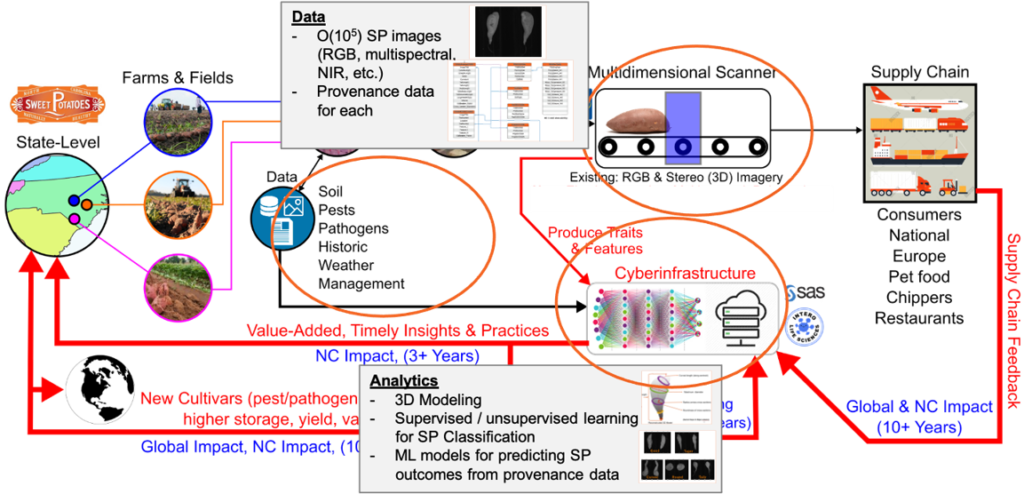
Project Description: The goal of this project is developing new data analysis tools for streamlining the North Carolina sweetpotato industry. A nexus point of all crop value is located at sorting facilities, where produce is sorted into value-added categories. These physical characteristics will be linked to up-stream provenance data (field location, weather data, management practices) and down-stream value (consumer preferences, storage life, etc.) to develop new management practices that maximize value. Currently, our lab is developing aspects related to the back-end machine learning classification techniques for quantifying sweetpotato phenotypes. For example, the EnBiSys Lab is developing a standard for data ingestion and pipeline access to generate knowledge graphs that allow visualization of data from the field to the table. This research is carried out in collaboration with the interdisciplinary research team funded by the Game-Changing Research Incentive Programs for the Plant Science Initiative (GRIP4PSI) here at NC State.
Contributors: Hangjin Liu, Azizah Conerly, Rashmi Datta, Srija Movva and Yash Sanjay Sonar
Molecular Mechanisms and Dynamics of Plant-Microbe Interactions at the Root-Soil Interface: InRoot
Project Description: The project is one of three projects funded by the Novo Nordisk Foundation under the Collaborative Crop Resilience Program. This is an international collaboration between NC State, The University of Copenhagen, Aarhus University and the Technical University of Denmark. The goal of these projects is to take a systems-level view of plants and their interactions with microbes in the soil, roots and foliage to improve plant health and productivity. This is particularly important in the face of climate change and emerging pathogens and pests where leveraging microbes to help plants avoid stresses while acquiring nutrients to reduce fertilizer, pesticide and irrigation become important. Our lab is developing multiscale models across biological scales that can predict the contribution of key plant and microbe interactions, which mediate intra- and inter-organismic resource allocation and promote plant fitness and resilience.
Contributors: Kirtley Amos and Max Gordon
Convergence Informatics and Data Access for the Science and Technologies for Phosphorus Sustainability (STEPS) Center
Project Description: The STEPS Center is an NSF Science and Technology Center focused on phosphorus sustainability through the integration of research across disciplines. As one of STEPS research initiatives, convergence informatics seeks to bridge the gaps between their research projects using integrative data management coupled with data-driven models and ML-based approaches. The EnBiSys Lab is contributing to STEPS convergence informatics by developing knowledge graphs as a visual representation of STEPS data, where these knowledge graphs capture how data is connected across multiple scales and projects. The goal is to create a dynamic, user-friendly website for STEPs researchers to search, compare and visualize the data collected across STEPS projects and disciplines.
Contributors: Azizah Conerly
Automating the Assessment of Severity of Southern Leaf Blight (SLB) in Corn using Machine Learning
Project Description: Biotic stresses, such as SLB, may visually appear on a crop and allow for the quantification of infected regions. The quantification of such stress, which includes the scoring of plants based on the proportion of infected areas to healthy tissue, is often performed manually based on visual symptoms, and is thus, time intensive, causing crop loss due to a delay between disease identification and management efforts. The EnBiSys Lab is taking two parallel approaches to automate the detection and grading of SLB in corn, which allows for disease detection at the plant and the plot levels. The first approach is focused on using computer vision and machine learning to perform quantification and prediction of biotic stress in near isogenic lines of corn plants in an effort to develop a consistent and objective method to score leaves efficiently. The second approach focuses on using hyperspectral images of whole fields of corn to detect and grade areas affected by SLB.
Contributors: Grace Vincent and Chanae Ottley
Soybean Grower Decision Support Tool
Project Description: To ensure continued on-farm profitability, growers must have access to dynamic tools that allow them to use data science to advance the economic and environmental sustainability of their operations. The EnBiSys Lab is leading the development of a web-based grower decision support dashboard that will be easily navigable by growers across North Carolina. Using SAS Viya, a cloud-based Al tool for analytics and data management, and machine learning, this dashboard is being created using field data generated since 2019 that comprises 50,000 data points on the relationship of production practices with yield and quality across the state. The goal of this project is to ultimately develop web-accessible models that provide data-driven solutions about soybean production practices directly to growers.
Contributors: Somshubhra Roy
Application of Deep Learning Algorithms for Multiple Environment Soybean Performance Predictions
Project Description: In collaboration with BASF, this project aims to predict soybean yield production from genotype, phenotype, and environmental data. The EnBiSys lab uses state-of-the-art deep learning algorithms that will provide efficient and explainable prediction results to provide optimal planting solutions for BASF.
Contributors: Peiran Wang
Systematically Exploring the Regulation of Gene Expression in Maize
Project Description: In collaboration with the Nelms Lab at the University of Georgia, high throughput screening (HTS) techniques are used to ectopically express transcription factors (TF) in Maize protoplasts. RNA-seq data is collected to measure gene expression in cells and to gain insights into the mechanisms of gene regulation. The EnBiSys Lab is leading the development of an improved statistical framework to analyze complex gene expression data to overcome technical and biological variations that are typical of HTS protocols. The lab is also taking the lead in deconvolving genetic regulatory networks from the RNA-seq data to uncover cellular programming and produce new understandings of the function of specific TFs. These experiments will offer a greater understanding of TF function and aid in engineering plant systems. This project aims to accelerate plant breeding to create the ‘crops of tomorrow,’ which is increasingly important in a changing climate.
Contributors: Teague McCracken and Max Gordon
Exploring Translation Mechanisms Through Upstream Open Reading Frames in Plants
Project Description: The goal of this project is to analyze open reading frames (uORFs) within the 5’ leader sequences and explore their impact on translation efficiency. It aims to enhance our comprehension of the mechanisms influencing uORFs activity. The outcomes of this research are guiding decisions in genomic breeding. Currently, the EnBiSys lab is applying machine learning methods to characterize the properties associated with uORFs.
Contributors: Peiran Wang
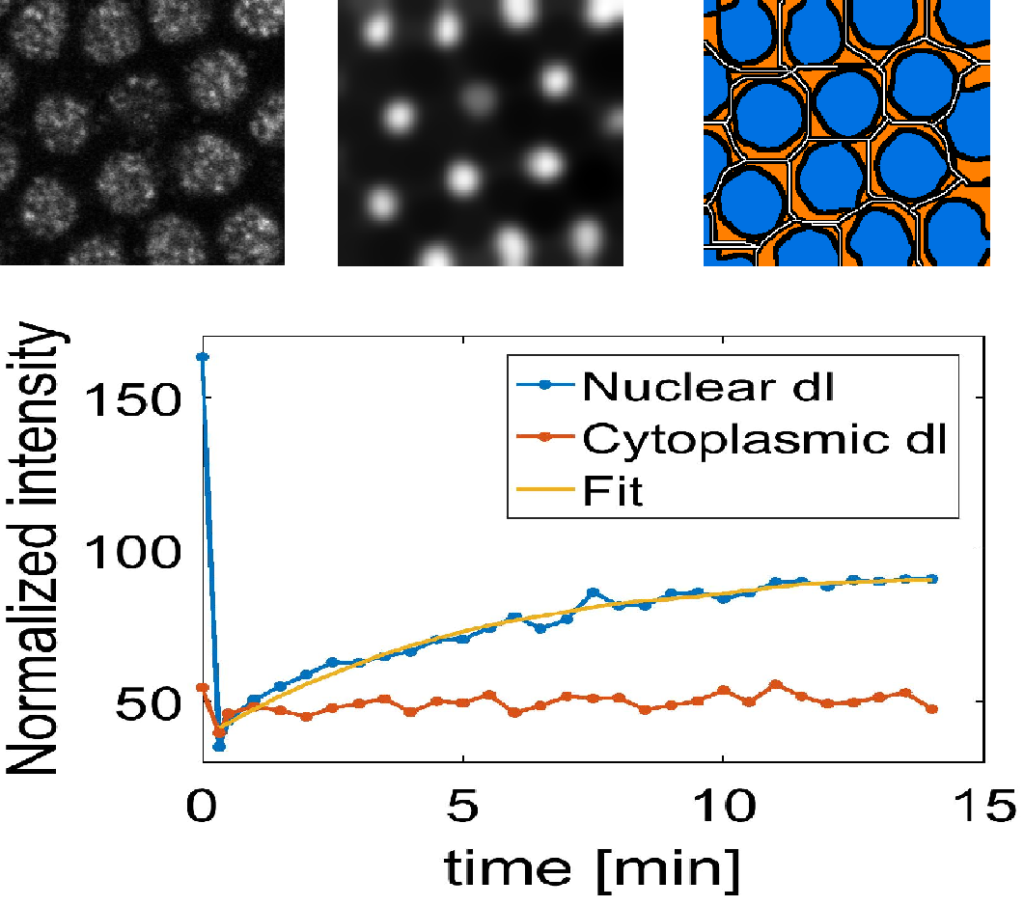
Systems-level measurements of biophysical parameters in the Dorsal/NF-kappaB pathway
Project Description: The Dorsal protein plays a vital role in the differentiation (patterning) of cells in Drosophila. The accurate measurement of the dorsal gradient is important to gain a better system level understanding of the developmental biology, signaling dynamics and gene regulation. The overall objective of this project is to perform detailed measurements of local biophysical parameters and global morphogen gradient properties to build and constrain a predictive, computational model of the Dorsal/NF-κB gradient in the early Drosophila embryo. The models generated as part of this project will test the central hypothesis of whether these models necessarily contain “sloppy parameters,” (wide ranges of estimated parameters that fit the experimental data) or may lead to discovering additional aspects that can improve the model. Our research also includes our general knowledge of the NF-κB signaling module that can be found in animals from Cnidarians to humans. Our lab is implementing computer vision and image processing techniques to quantify the spatiotemporal behavior of dorsal gradient and build predictive models using mathematical modelling of the parameters involved in this process. This research is performed in collaboration with the Reeves Lab at Texas A&M Univeristy.
Contributors: Sharva Hiremath